PROSPECTS
Covid-19 testing Working at the Glasgow Lighthouse Lab
Between graduating from her undergraduate degree in genetics at the University of Glasgow and embarking on her PhD, Holly Kerr worked in the Glasgow Lighthouse Lab as a laboratory scientist. Here she shares her experiences and takeaway lessons for anyone considering working in a biology laboratory

In February 2020, news was spreading that the World Health Organization (WHO) was about to declare Covid-19 a global pandemic. It was the morning of my interview at the Roslin Institute in Edinburgh for a PhD in one health and infectious disease, which was due to start in September 2020. I quickly added a picture of a test tube labelled ‘Coronavirus’ to my interview slides (see Figure 1). In the interview, I said something along the lines of ‘…and with news this morning that Covid-19 may gain pandemic status, infectious diseases are still a major and relevant threat to health across the globe, reinforcing the importance of the One Health agenda’. Little did I know that a few months later, the only reason I would be leaving the house was to handle and process similar-looking tubes in full PPE for hours on end.
Some pretty major events on both an individual and global scale happened following that interview. Most excitingly for me, I was offered and accepted a place on the PhD programme the next day. The next month (March 2020), the UK went into lockdown. I sat my final year undergraduate exams in my bedroom and held a ‘graduation ceremony’ with my mum and dog in the garden. By summer, I was working with one of the UK’s largest Covid-19 testing laboratories. It is fair to say that the year did not go as expected for most of us. However, I feel fortunate to have had valuable opportunities from an otherwise terrible time that are likely to shape my career path for years to come.

Joining the Lighthouse Lab
The Lighthouse Lab in Glasgow is one of five mass testing UK Biocentre laboratories set up from scratch in response to the Covid-19 outbreak. I joined in June 2020 as a laboratory scientist, one of two main roles available, depending on experience. I presumed my experience to be fairly limited as I had only just graduated with a BSc in genetics. Most early recruited employees were established researchers from laboratory groups within the university, working as research assistants, PhD students, post-docs, research associates, even principal investigators, who head their own research groups. I had imposter syndrome when I arrived. But my honours degree and other past experience meant I was well prepared for the work I was going to do.

I was familiar with a laboratory environment, thanks to the practical lab sessions and final year wet-lab project at university, as well as the 8-week lab internship I arranged in the summer before my final year. I was familiar with the experimental techniques the laboratory used to test for presence of the virus in a sample – quantitative polymerase chain reaction (qPCR, see Box 1). In genetics, PCR is our bread-and-butter method to test for the presence or absence of a gene of interest, for example, or to prepare DNA for sequencing. The year before the pandemic, I had volunteered with the Glasgow Science Centre’s outreach project to teach Higher (equivalent to A-level) biology students about the use of PCR and DNA sequencing to test for different viral infections. Never underestimate the value of the experience, skills and knowledge you gain throughout all stages of your education, work experience and volunteering. As I have learned, it can be highly relevant in the future.
Box 1 Using quantitative PCR to test for Covid-19
By the time you read this, you may have experienced the uncomfortable feeling of a swab reaching down to your tonsils and far up your nose, as part of a Covid-19 test. But what happens to the sample, and how do the scientists decide if your test is positive or negative for Covid-19? The answer lies in the polymerase chain reaction (PCR), developed in 1985 by Kary B. Mullis. PCR amplifies DNA in a sample to make billions and billions of copies. You can target the procedure to amplify whatever segment of DNA you are interested in, and you can make it quantitative by determining the original starting concentration of the sample. In testing for Covid-19, this means that we can take a sample and ask, is there viral genetic material here? And if so, how much?
A mixture of cells is obtained on the swab and collected in the liquid in the tube. The cells include some of your own cells, and some of the trillions of microbes that reside on every bit of your body. If you are infected with a virus, there is a high chance that some viral particles will be captured on that swab. Since SARS-Cov-2 genetic information is stored in RNA, it is first reverse-transcribed to make complementary DNA that the PCR test can work on.
Strings of A, C, T and G nucleotides called primers are designed to match the ends of known Covid-19 genes. If the Covid-19 virus is in the sample, the primers will attach and amplify these genes in a chain reaction (see Figure 2.1). This means that even extremely small amounts of starting virus can be detected and a positive result is then determined.
Swab to test result: a finely tuned production line
However, I was not prepared for the scale of qPCR that was required for mass testing. The laboratory was housed over several floors of the University of Glasgow Teaching and Learning building. Most of the work happened in different rooms along a long corridor. Each room is kitted out with multiple versions of the same equipment required for each step of the process, known as the workstations. I was initially involved with the first step, called ‘unbagging’. Samples arrive in large boxes and must be carefully taken out of their double bags, checked for leakages, and racked before the next step.
At this stage, any sample containing the virus is still potentially infectious, so we worked under a safety airflow hood with full personal protective equipment (PPE): mask, visor, gloves taped on, gown, apron, sleeves, more gloves. The hum of the hoods that filled the laboratory was masked slightly by our background music. Sample numbers soon climbed to 10000, 20000 and then 35000 a day. More employees were recruited, and the laboratory began to operate around the clock on two shifts. Those with laboratory experience were moved to the workstations after initial sample unbagging, which became the role of sample handlers.
In order to get through the vast truckloads of samples that arrived day and night, the laboratory had to operate like a smooth, well-oiled machine. As of February 2021, the Lighthouse Lab network’s capacity reached over 700 000, more than the entire population of Glasgow in a single day.
My main role was then in the scanning workstation. The samples have to be carefully tracked and associated with a barcode on the laboratory’s computer system so that each person gets their own result. These initial steps are really important before the qPCR to determine a positive or negative result can take place.
Eventually, the laboratory implemented automation. Robotic machines scan the barcodes in a fraction of the time it took us mere humans. The robots also replaced much of the manual pipetting that was needed. Although at first they were not perfect, the robots resolve much of the human error in the process. Operating and optimising the first robots was a great experience, and one not available in most research laboratories.

Targets and teamwork
To be part of those early stages was daunting, but exciting. The laboratory was processing more and more tests and getting the test results out more quickly with every shift. Since the work is highly repetitive, having targets, milestones and friendly competition in the lab is crucial. We were driven largely by numbers and statistics, with a goal to process as many test results accurately in the quickest turnaround time possible. There was no lack of motivation – the world had stopped due to the virus and every member of staff was affected by Covid-19 in some way. Quality, as well as quantity, was important too. Every sample was a person at home waiting on their result, and we treated them as such.
We worked to standard operating procedures (SOPs). Non-conformity forms (concerning SOPs that were not achieved) were carefully reported and discussed every week. The SOPs were constantly scrutinised and updated and after a few months of the laboratory opening, a dedicated quality-assurance team was established. Any member of staff could suggest changes that would make a step in the process more efficient. Everyone played a role, and everyone had a say.
As I settled into the laboratory, my imposter syndrome faded. The hierarchy that often exists in a research environment was largely absent in the Lighthouse Lab. There were different roles, but each was equally important in the whole operation. Working directly with, and learning from, experienced researchers without the feeling of pestering them or being somehow ‘below’ them in a chain of importance felt like an incredible opportunity. In a room full of people, being the least experienced means you have the best chance to learn.
Box 2 The swab-to-result process of a Covid-19 test using quantitative PCR
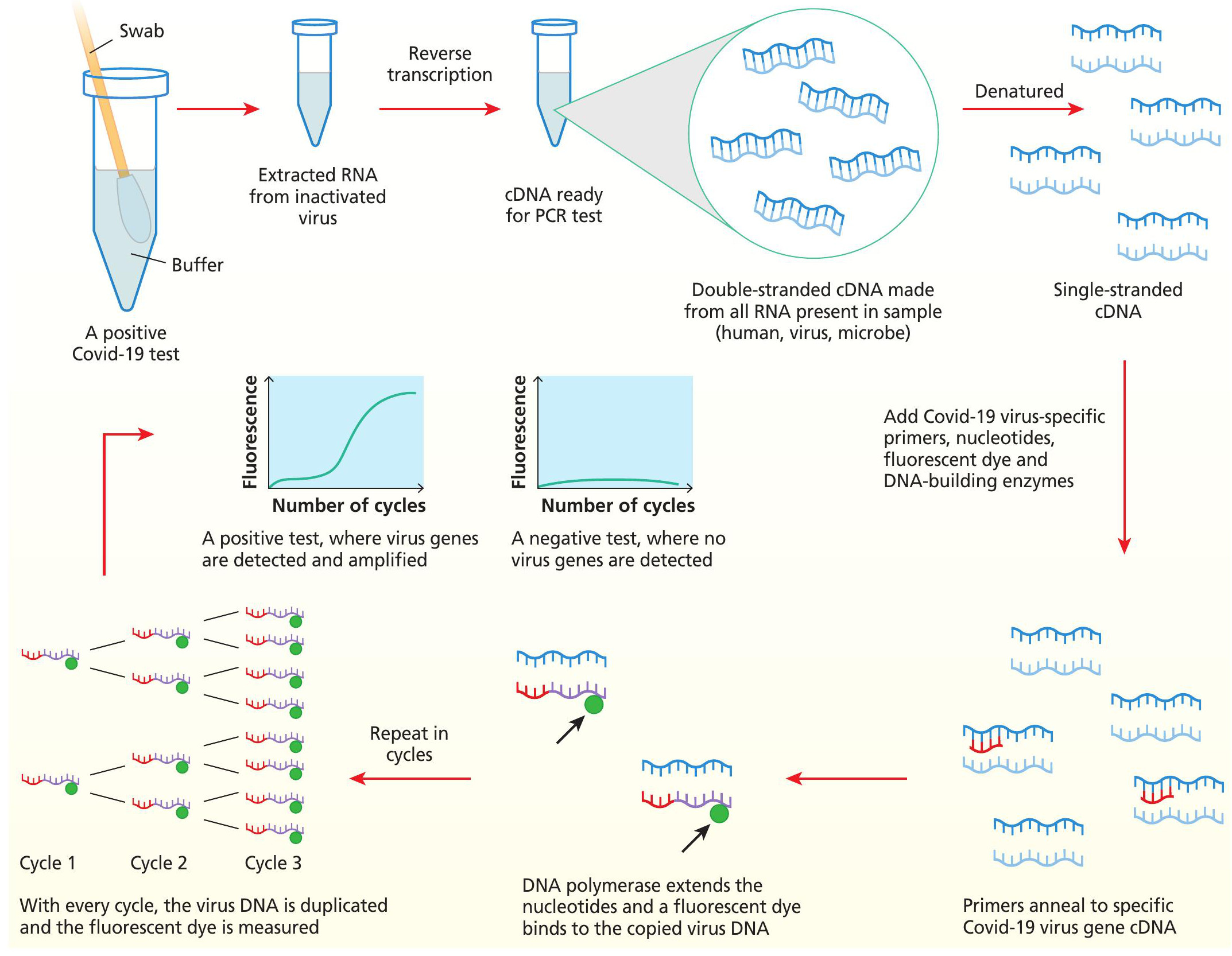
A nose and throat swab is put in a buffer and taken to the testing laboratory, where any potential virus in the tube is first inactivated and then RNA is extracted. For the qPCR test, the RNA is reverse transcribed to complementary DNA (cDNA). cDNA is denatured to expose open single-stranded cDNA which the Covid-19 gene specific primers can bind to. DNA polymerase extends the strings of nucleotides where primers have matched to Covid-19 genes in the sample and this releases a fluorescent dye. The process is repeated in multiple cycles so that in a positive test, the virus DNA is duplicated every time and fluorescence can be detected in greater amounts every cycle. In a negative test, there is no original RNA, so no Covid-19 gene cDNA for the primers to bind and thus, no fluorescence can be detected.
My few months at the Lighthouse Lab in its early stages gave me a great insight into a job in a science laboratory. It taught me to appreciate the value of my past experience and gave me a real chance to be part of a large, functioning team. It has been a stark contrast to the start of my PhD, which has mostly involved working in isolation. Nonetheless, I now have the opportunity to research the virus itself and work with an even bigger international, multidisciplinary team at the university. Exploring different working environments will help me to decide in future my preference for a long-term career. It is great to know exactly what you want to do in the future but in reality most of us are not sure. Dipping your toes in different jobs and career paths is a good way to find out. Always appreciate the value in all your experiences and make the most of the opportunities that come your way.
TERMS EXPLAINED
Imposter syndrome A feeling that you are not as competent as people think you are.
Quantitative polymerase chain reaction (qPCR) A method used to measure the presence and abundance of a target section of DNA present in a sample.
Standard operating procedures (SOPs) A document detailing the step-by-step instructions for regular procedures within a laboratory, so that the procedure is always carried out correctly and in the same manner.





